A novel
approach to predict protein backbone conformation
There are
two server operation modes:
|
Predict Local Structure Classes |
actually
predicting structural classes by sequence. The amino
acid sequence is fed to the input. |
|
Compare prediction for Known Structure |
is a
prediction quality analysis for a known protein chain
structure. A PDB file is input. |
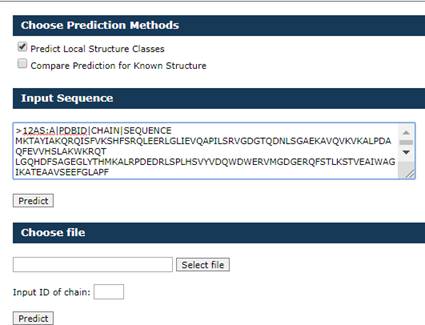
Input for Predict Local Structure Classes mode
Provide in
the textbox protein sequences in free FASTA format as shown below.
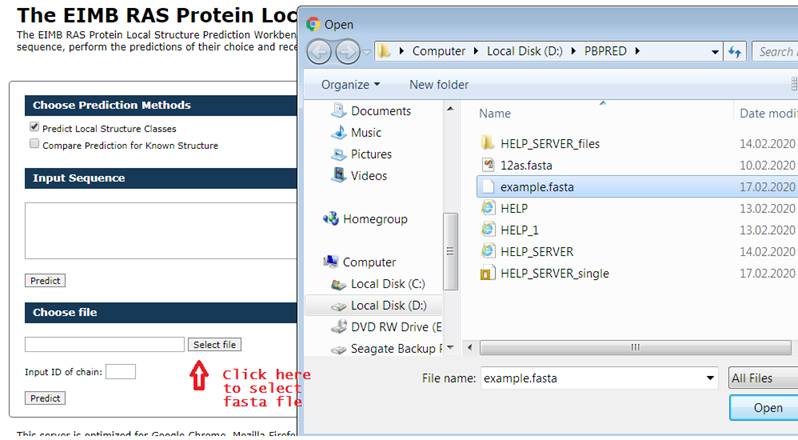
Another way
to enter the amino acid sequence is to download the FASTA file.
Below is an
example screenshot.
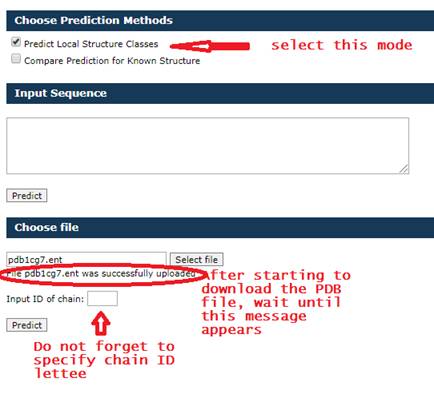
Input for Compare prediction for Known Structure mode
You must select a PBD file and do not forget to select the
ID of chain
Than click
the predict button and wait approximately 10 seconds.
An example
of filling out the form is shown below.
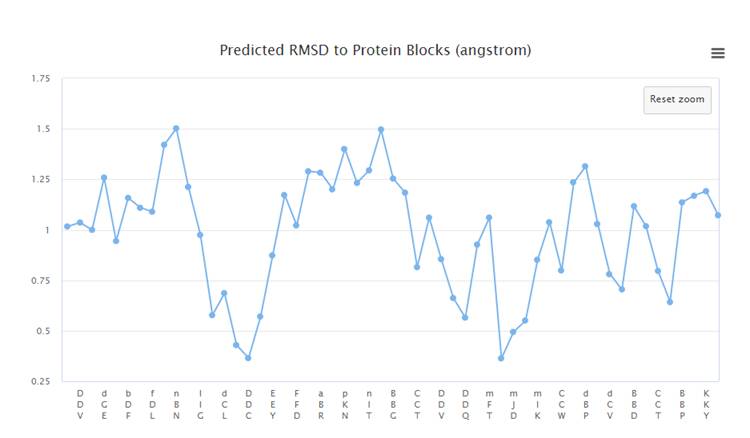
Results
The
presentation of the results for the Predict Local Structure Classes mode and
Compare prediction for Known Structure
mode is
almost the same. The difference is
that in the second case, you can compare the predicted classes of the
local
structure with the data from the PDB file. The value Q16 is calculated - the
percentage of correct predictions for 16 classes
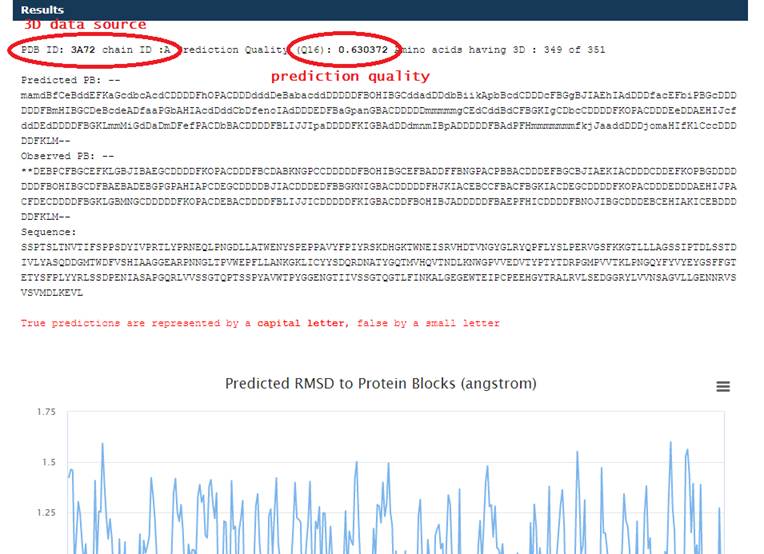
On the
graph, you can select and enlarge the area of interest.
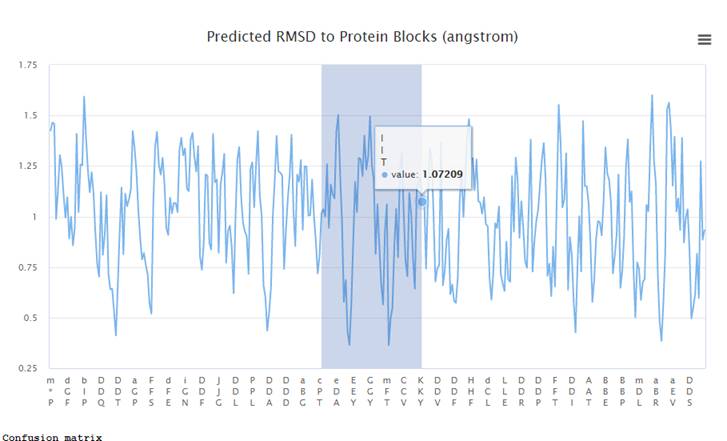
Below is a
graph of the area highlighted in the previous graph